Our research
The Natarajan laboratory We are interested in the fundamental process of cell fate specification during mammalian development and differentiation at single-cell level. The group aims to understand general paradigms of cell state and fate specification, with a particular interest in transcriptional and epigenetic regulatory networks, and their crosstalk with cell cycle, metabolism and fundamental cellular processes.
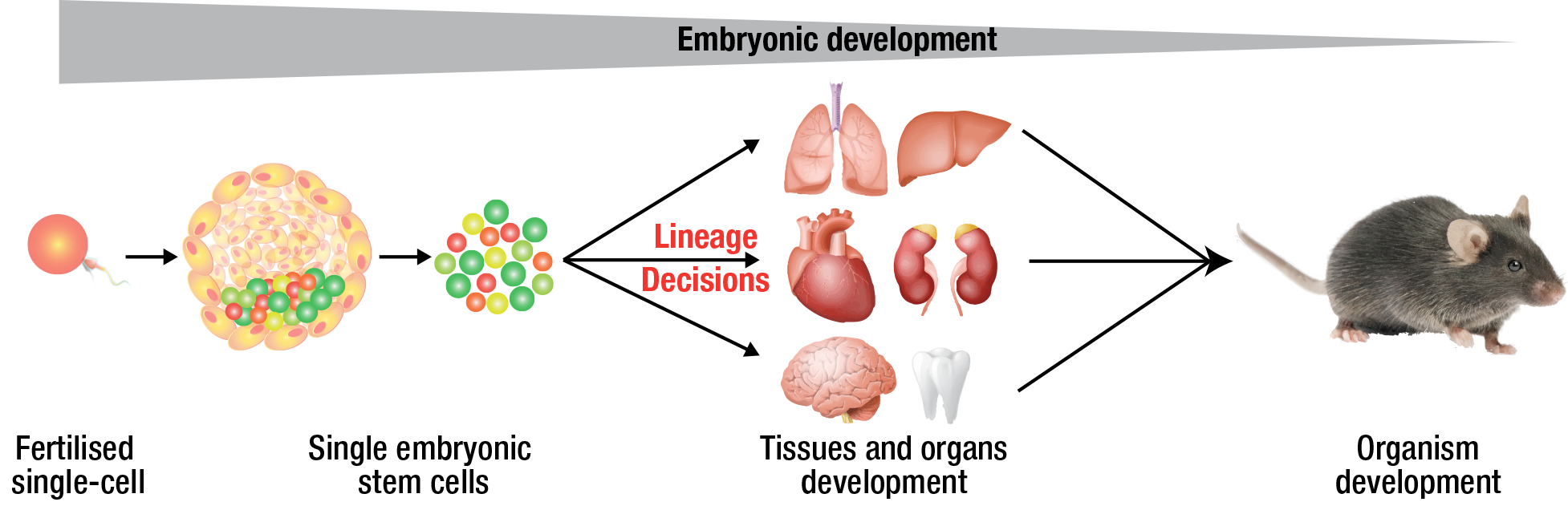
We have a interdisciplinary foundation and combine wet-lab experiments with theoretical models and computational approaches at single-cell and bulk level to uncover principles governing cell fate decisions, and their consequences in development and disease. We leverage different developmental (ESCs/hiPSCs), differentiation (CNS, immune) models alongside molecular, cellular and computational approaches.
Areas of research
Transcriptional and Epigenetic control of iPSC differentiation to CNS cell types
We study transcriptional and epigenetic crosstalk during iPSC differentiation to CNS cell types (microglia, neurons) via single-cell omics profiling and computational biology.
Cell fate specification and dynamics during differentiation
We develop and apply causal gene regulatory network inference to study factors, interactions, pathways driving cell-state and fate-specification from multi-modal single-cell datasets
New methods for single-cell biology
We have a strong interest in development, extension and utilisation of new experimental and computational methods for single-cell biology.
Working closely with collaborators, we develop new computational tools that integrate measurements from several single-cell technologies.
Our Team

Kedar Natarajan
Group Leader

Malvika Sudhakar
Postdoctoral Fellow

Gergo Kovacs
Postdoctoral Fellow

Yee Ying Lim
PhD Student

Alvaro Bustos Gutierrez
PhD Student

Harie Vignesh
Research Assistant

William Echwald
Masters thesis student

Victoria Nicolaysen
Shared lab technician
Join us!
We welcome enquiries from highly interdisciplinary, motivated and ambitious candidates.
Postdocs candidates with strong background and interest in areas of single-cell genomics, computational biology and machine learning are welcome to enquire informally on lab projects. We encourage and support applications to Danish and international funding agencies. Please email a tailored cover letter, CV, publication record and details of two references
Interested PhD students are also welcome to enquire about applying for funding. Please email directly.
Publications
2024
Irastorza-Azcarate I, Kukalev A, Kempfer R, Thieme CJ, Mastrobuoni G, Markowski J, Loof G, Sparks TM, Brookes E, Natarajan KN, Sauer S, Fisher AG, Nicodemi M, Ren B, Schwarz RF, Kempa S and Pombo A
Preprint in: BiorXiv
2023
Vlassis A, Jensen TL, Mohr M, Jedrzejczyk DJ, Meng X, Kovacs G, Gómez MM, Barghetti A, Sergi M Abad SM, Baumgartner RF, Natarajan KN, Nielsen LK, Warnecke T, Gill RT
Published in: iScience
Petrosius V, Aragon-Fernandez P, Üresin N, Kovacs G, Phlairaharn T, Furtwängler B, Beeck JOD, Skovbakke SL, Goletz S, Thomsen SF, Keller UAD, Natarajan KN, Bo T. Porse & Schoof E
Published in: Nature Communications
Lee J, Møller AF, Chae S, Bussek A, Park TJ, Kim Y, Lee HS, Pers TH, Kwon T, Sedzinski J, Natarajan KN
Published in: Science Advances
Agrawal R and Natarajan KN
Published in: Advances in Cancer Research
Mishra S, Pandey N, Chawla S, Sharma M, Chandra O, Jha IP, SenGupta D, Natarajan KN# Kumar V#
Published in: Genome Research
G Weng, J Kim, KN Natarajan#, KJ Won#
Preprint in: bioRxiv
2022
Madan E, Palma AM, Vudatha V, Trevino JG, Natarajan KN, Winn RA, Won KJ, Graham TA, Drapkin R, McDonald SAC, Fisher PB, Gogna R
Published in: Cancer Research
Madsen MS, Broekema MF, Madsen MR, Koppen A, Borgman A, Gräwe C, Thomsen EGK, Westland D, Kranendonk MEG, Koerkamp MG, Hamers N, Bonvin AMJJ, Pittol JMR, Natarajan KN, Kersten S, Holstege FCP, Monajemi H, van Mil SWC, Vermeulen M, Kragelund BB, Cassiman D, Mandrup S, Kalkhoven E
Published in: Nature Communication
2021
Kim J, Jakobsen ST, Natarajan KN# and Won KJ#
Published in: Nucleic Acid Research.
2020
Moeller AF and Natarajan KN#
Published in: Life Science Alliance.
Keating SM, Waltemath D, et al., and SBML Level 3 Community members
Published in: Molecular Systems Biology.
Terkelsen MK, Bendixen SM, Hansen D, Scott EAH, Moeller AF,
Nielsen R, Mandrup S, Schlosser A, Andersen TL, Sorensen GL, Krag A,
Natarajan KN, Detlefsen S, Dimke H, Ravnskjaer K
Published in: Hepatology.
Cuomo SE, Seaton DD, McCarthy DJ, Martinez I, Bonder MJ,
Garcia-Bernardo J, Amatya S, Madrigal P, Isaacson A, Buettner F, Knights A,
Natarajan KN, HipSci Consortium, Vallier L, Marioni JC, Chhatriwala M, Stegle O.
Published in: Nature Communications.
Oatley M, Bolukbasi OV, Svensson V, Shvartsman M, Ganter K,
Zirngibl K, Pavlovich PV, Milchevskaya V, Foteva V, Natarajan KN, Baying B, Benes V,
Patil KR, Teichmann SA, Lancrin C
Published in: Nature Communications.
2019
Howick VM, Russell AJC, Andrews T, Heaton H, Reid AJ, Natarajan K,
Butungi H, Metcalf T, Verzier LH, Rayner JC, Berriman M, Herren JK, Billker O, Hemberg M,
Talman AM, Lawniczak MKN.
Published in: Science.
Natarajan KN*#, Miao Z*, Jiang M*, Huang X, Zhou H, Xie J, Wang C, Qin S,
Zhao Z, Wu L, Yang N, Li B, Hou Y, Liu S, Teichmann SA#.
Published in: Genome Biology.
Natarajan KN#.
Published in: Methods in Molecular Biology.
2018
Chen X, Miragaia RJ, Natarajan KN, Teichmann SA.
Published in: Nature Communications.
Pramanik J, Chen X, Kar G, Henriksson J,
Gomes T, Park JE, Natarajan K, Meyer KB, Miao Z, McKenzie ANJ,
Mahata B, Teichmann SA.
Published in: Genome Medicine.
Tuck AC, Natarajan KN, Rice GM, Borawski J, Mohn F, Rankova A, Flemr M, Wenger A,
Nutiu R, Teichmann SA, Bühler M
Published in: Life Science Alliance.
2017
Natarajan KN*#, Teichmann SA, Kolodziejczyk AA.
Published in: Current Opinion of Genetics and Development.
Kar G, Kim JK, Kolodziejczyk AA, Natarajan KN, Torlai Triglia E, Mifsud B, Elderkin S, Marioni JC, Pombo A, Teichmann SA
Published in: Nature Communications.
Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M.
Published in: Nature Methods.
Natarajan KN*# and Teichmann SA (Journalistic feature)
Published in: Nature methods.
Natarajan KN*# (Special feature comment)
Published in: Cell.
Svensson V*, Natarajan KN*, Ly LH, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A, Teichmann SA.
Published in: Nature Methods.
2016
Proserpio V, Piccolo A, Haim-Vilmovsky L, Kar G, Lönnberg T, Svensson V, Pramanik J,
Natarajan KN, Zhai W, Zhang X, Donati G, Kayikci M, Kotar J, McKenzie AN, Montandon R, Billker O, Woodhouse S, Cicuta P, Nicodemi M, Teichmann SA.
Published in: Genome Biology.
Rodriguez N, Pettit JB, Dalle Pezze P, Li L, Henry A, van Iersel MP,
Jalowicki G, Kutmon M, Natarajan KN, Tolnay D, Stefan MI, Evelo CT, Le Novère N.
Published in: BMC Biology.
2015
Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J,
Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, Marioni JC, Teichmann SA
Published in: Cell Stem Cell.
Scialdone A, Natarajan KN, Saraiva LR, Proserpio V, Teichmann SA, Stegle O, Marioni JC, Buettner F
Published in: Methods.
Buettner F*, Natarajan KN*, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O.
Published in: Nature Biotechnology.
2014
Chelliah V, Juty N, Ajmera I, Ali R, Dumousseau M, Glont M, Hucka M, Jalowicki G,
Keating S, Knight-Schrijver V, Lloret-Villas A, Natarajan KN, Pettit JB, Rodriguez N, Schubert M,
Wimalaratne SM, Zhao Y, Hermjakob H, Le Novère N, Laibe C
Published in: Nucleic Acid Research.
2011
Jalowicki G, Rodriguez N, Kutmon M, Pettit JB, Li L, Henry A, Natarajan KN, Laibe C, Evelo CT, Novere NL
PrePrint in: Nature Precedings.
Funding
Our research is possible due to generous support form:



Contact Us
Kedar Natarajan is Associate Professor at DTU Bioengineering at Technical University, Denmark.
The group (Transcriptional regulation and Single-cell systems genomics) is a part of DTU Bioengineering
at Technical University of Denmark.
Transcriptional regulation and Single-cell systems genomics,
DTU Bioengineering
Technical University of Denmark
Søltofts Plads, DK-2800 Kgs. Lyngby
reachknn@gmail.com